itcrispr — An interface to assemble and track progress of CRISPR sequence experiments in BioNumerics¶
Version: 0.1.4
About¶
A program to assist in the analysis of CRISPR sequence experiments in BioNumerics. Assembly is done using predefined primers. The finished status of each primer is saved within the program.
Setup and configuration¶
Install Python 2.6¶
Download (x86 version): http://www.python.org/download/releases/2.6.6/
Install PyQt¶
Download (x86 version for Python 2.6): http://www.riverbankcomputing.co.uk/software/pyqt/download
Install itcrispr¶
Clone repository
git clone git@github.com:vkvn/bionumerics_scripts.git
Build and install
cd bionumerics_scripts/itcrispr
python setup.py build
python setup.py install
Configure script¶
Change configuration in
C:\Python26\Lib\site-packages\itcrispr\itcrispr.cfgDATABASES,EXPERIMENTSandPRIMER_PATHare all required for the program to work.DATABASESThe database name as in BioNumerics. Multiple databases can be specified as comma-separated values. This option is to prevent the script from being executed accidentally in other databases.
EXPERIMENTSList of CRISPR sequence experiments - comma-separated.
PRIMER_PATHPATH to the directory containing primer files in text format. File extension should be
.txtand file should contain only the sequence (FASTA and other formats not supported).
Usage¶
Select entries in BioNumerics. If no selection is made, program runs through all entries in the database.
Run script using the
Script --> Run script from fileoption in BioNumerics. Navigate to theC:\Python26\Scriptsfolder and selectrun_itcrispr.pyNote
As easier method to run the script would be to add a button to the toolbar. Steps for doing this is described in Add a button to the toolbar.
An assembly will be done using the CRISPR sequences and primers. A dialog window will then be displayed with the list of primers that aligned with the sequences. Double-click on a primer to navigate to that region in the assembler.
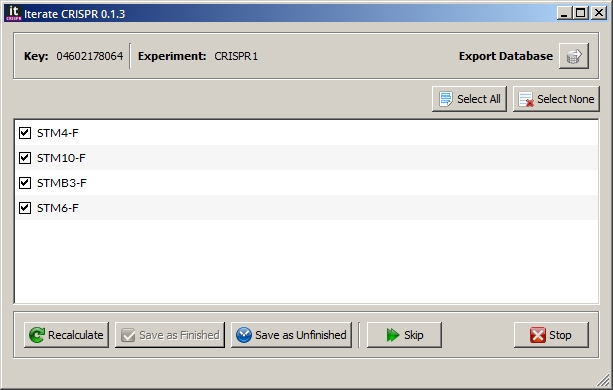
If you consider an experiment for an entry as finished, click the Save as Finished button. No primer should be checked. Use the Select None button to deselect all primers.
To save an experiment as unfinished, check the primers for regions that need to be repeated and click the Save as Unfinished button.
If you made some changes in the assembler window, use the Recalculate button to redo the assembly.
Use the Skip button to redo this entry at a later time.
Use the Export Database button to export the entire database to a
CSVfile.Stop button will terminate the program.
Notes¶
- If the experiment for an entry is saved as finished, it will not be displayed in subsequent runs.
- Primer sequences are read, an assembly is done after changing the minimum score to the smallest length of primer.
- Primers selected previously appear checked when the window is shown.
- Closing the window skips to the next experiment. Use the Stop button to exit the program.
- Sequence approval status in BioNumerics is not taken into consideration for deciding which entries to check. Sequences are made approved if they are saved as finished using the program.
Add a button to the toolbar¶
This needs to be done whenever the program is updated!
Copy
C:\Python26\Lib\site-packages\itcrispr\itcrispr-menu.bnstothe scripts home directory under your BioNumerics Home directory. For example,
C:\Users\vimal\BioNumerics\Data\Scripts Home\Load this file in the BNS Script editor
Script --> BNS Script editorfollowed by
File --> Load ScriptOpen
File --> Attachmentsand click Add newSelect
C:\Python26\Scripts\run_itcrispr.pyand click Open. Enterrun_itcrisprin the dialog that appears and click OK.Save the script.
Restart BioNumerics. An icon
 should now be available in the toolbar. Click
this icon to launch the program.
should now be available in the toolbar. Click
this icon to launch the program.
Add or remove primer sequences¶
Primer files are plain text files with just the sequence of the primer.
Add or remove primers to/from the location specified in the configuration
file - the PRIMER_PATH variable.